cztpipelineIn this section, some illustrative examples of usage of cztpipeline for a few specific cases are shown.
To view the help of cztpipeline, use cztpipeline -h or czthelp cztpipeline To get information on a specific input keyword, use cztpipeline -h keyword.
In this example, spectrum of a source for the period of entire observation will be generated starting from level-2 data archive. For this, we consider an observation of crab with observation id 9000002432. As the first step, download the level-2 data zip file from Astrobrowse for this observation and then unzip the same as follows at desired location
$ unzip LEVEL2AS1CZT20181014C04_007T02_9000002432.zip
This would generate a directory named 20181014_C04_007T02_9000002432_level2 which contains the standard level-2 products including spectra. However, it is likely that the products are generated with previous versions of software and CALDB. Still, there would not be any changes in the products until end of stage-1 of the data analysis pipeline. Hence, we will execute the cztpipeline stages 2 and 3 using the level-2 products files from stage 1 that was available in the downloaded data.
As our objective is to generate spectrum for entire observation, the inputs to cztpipeline are straightforward as shown in the example below. Note that the level-2 directory given as the input is where the output files will also be generated and will replace the original files in the example given below.
$ cztpipeline
--------------------------------------------------------------------------------------------------
ASTROSAT CZTI PIPELINE
Version: V3.0 December 2022
--------------------------------------------------------------------------------------------------
Enter Start Step [2]: 2
Enter End Step [3]: 3
Enter path to create L2 pipeline products []:
/home/user/path/data_czti/20181014_C04_007T02_9000002432_level2
Enter full path of user-gtifile [-]:
Enter output type <lc/spec/both>(for cztbindata) [both]: spec
Apply maskweights (YES/NO)(for cztbindata) [Yes]: Yes
Enter RA of source in decimal degrees [0]: 83.63308
Enter DEC of source in decimal degrees [0]: 22.01446
Overwrite existing file?(YES/NO): [YES]: Yes
--------------------------------------------------------------------------------------------------
CZTPIPELINE PARAMETERS
--------------------------------------------------------------------------------------------------
Task Name : cztpipeline.py
Level2 Directory : /home/user/path/data_czti/20181014_C04_007T02_9000002432_level2
Entry Stage : 2
Exit Stage : 3
Right Ascension of Source : 83.63308
Declination of Source : 22.01446
User GTI File : -
Event Selection Algorithm : default
Clobber : YES
--------------------------------------------------------------------------------------------------
--------------------------------------------------------------------------------------------------
Do you want to continue? [Y/n]: Y
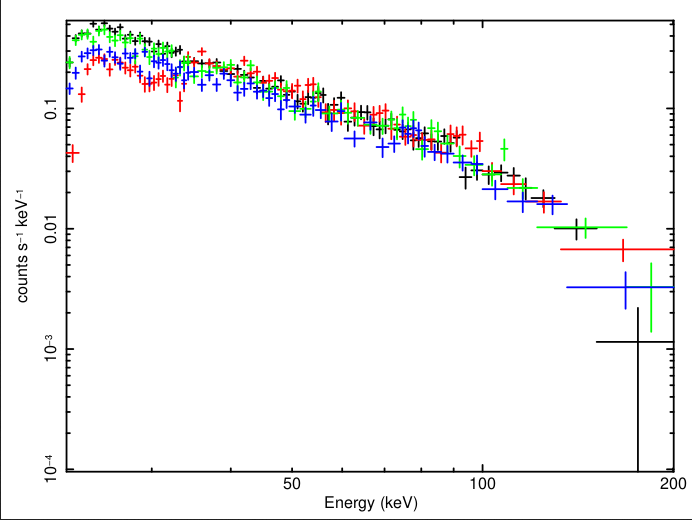
After this, cztpipeline would execute the modules of the pipeline and print the details on screen finally showing the message that the excution completed successfully. Spectral files and respective response files for each quadrant are generated within the level-2 directory under modeM0. Figure 1 shows the spectra of each quadrant for this observation loaded in XSPEC with below commands.
XSPEC12>data 1:1 AS1C04_007T02_9000002432cztM0_level2_quad_clean_Q0.pha
XSPEC12>data 2:2 AS1C04_007T02_9000002432cztM0_level2_quad_clean_Q1.pha
XSPEC12>data 3:3 AS1C04_007T02_9000002432cztM0_level2_quad_clean_Q2.pha
XSPEC12>data 4:4 AS1C04_007T02_9000002432cztM0_level2_quad_clean_Q3.pha
XSPEC12>ignore bad
 |
|---|
| Figure 1: CZTI crab spectra for all quadrants for obsid 2432 generated with cztpipeline and loaded in XSPEC. |
It is also possible to have optional suffix added to the output files genearted so that they have different names than that available in the level-2 data archive. For example, running cztpipeline with options as below would produce output files from stage 2 and 3 with a suffix ‘fullobs’ added to the file names.
$ cztpipeline suffix='fullobs'
--------------------------------------------------------------------------------------------------
ASTROSAT CZTI PIPELINE
Version: V3.0 December 2022
--------------------------------------------------------------------------------------------------
Enter Start Step [2] : 2
Enter End Step [3] : 3
Enter path to create L2 pipeline products [] :
/home/user/path/data_czti/20181014_C04_007T02_9000002432_level2
Enter full path of user-gtifile [-] :
Enter output type <lc/spec/both>(for cztbindata) [spec] :
Apply maskweights (YES/NO)(for cztbindata) [Yes] :
Enter RA of source in decimal degrees [83.63308] :
Enter DEC of source in decimal degrees [22.01446] :
Overwrite existing file?(YES/NO): [YES] :
This option of suffix to the file names will be useful when a user need to generate spectra for multiple intervals of an observation as discussed in Section 3.
For bright sources such as crab in the example above, it is always advisable to load quadrant wise spectra and fit them simultaneously allowing for cross-normlization constants. However, for faint sources (typically less than 100 mCrab), it may be useful to co-add spectra from all quadrants to get the total spectrum. CZTI data analysis pipline includes a task cztaddspec for this purpose. As an example, here we add quadrant-wise spectra generated from the previous example to get the total spectrum.
As cztaddspec requires a text file with list of spectrum files, first we create such a file named ‘speclist.txt’ with content as below:
AS1C04_007T02_9000002432cztM0_level2_quad_clean_Q0.pha
AS1C04_007T02_9000002432cztM0_level2_quad_clean_Q1.pha
AS1C04_007T02_9000002432cztM0_level2_quad_clean_Q2.pha
AS1C04_007T02_9000002432cztM0_level2_quad_clean_Q3.pha
Now execute cztaddspec from the directory containing these spectra as follows:
~/path/data_czti/20181014_C04_007T02_9000002432_level2/czti/modeM0$ cztaddspec
Enter Input List of spectrum files [] : speclist.txt
Enter Name of output spectrum file [] : AS1C04_007T02_9000002432cztM0_level2_quad_clean_quadadded.pha
Whether to add response files? [y] : y
Whether to add exposure times (same detector multiple obs)? [y] : n
[ cztaddspec.cpp:96] ----------------------------------------------------------------------------
[ cztaddspec.cpp:97] CZTADDSPEC PARAMETERS
[ cztaddspec.cpp:98] ----------------------------------------------------------------------------
[ cztaddspec.cpp:99] Modulename : cztaddspec_v3.0
[ cztaddspec.cpp:100] Input List file : speclist.txt
[ cztaddspec.cpp:101] Output spectrum file : AS1C04_007T02_9000002432cztM0_level2_quad_clean_quadadded.pha
[ cztaddspec.cpp:102] Output response file : AS1C04_007T02_9000002432cztM0_level2_quad_clean_quadadded.rsp
[ cztaddspec.cpp:104] Addresp : YES
[ cztaddspec.cpp:111] Addexposure : NO
[ cztaddspec.cpp:114] Clobber : YES
[ cztaddspec.cpp:118] History : YES
[ cztaddspec.cpp:121] ----------------------------------------------------------------------------
[ main.cpp:38] CZTADDSPEC STARTED....................
[ cztaddspec.cpp:146] Adding 4 spectra together
[ utils.cpp:1605] Template file used : /home/user/work/czti/gitczti/czti/czti_pipeline/czti/templates/spectrumTemplate
[ cztHeaderParam.cpp:314] Read all the header keywords
[ cztHeaderParam.cpp:531] Copied all the header keywords sucessfully
[ level2validation.cpp:4925] Total signal to noise ratio is 115.548
[ level2validation.cpp:4926] Selected good channels are 34 - 390
[ level2validation.cpp:4937] Grouping spectrum into 100 logarithmic bins
[ utils.h:732] Updating keys in file: AS1C04_007T02_9000002432cztM0_level2_quad_clean_quadadded.pha
[ utils.cpp:1605] Template file used : /home/user/work/czti/gitczti/czti/czti_pipeline/czti/templates/rspTemplate
[ main.cpp:49] CZTADDSPEC COMPLETED SUCCESSFULLY
[ main.cpp:51] Time Elapsed:0 seconds
This produces total spectrum file and corresponding response file. It may be noted that the same tool maybe used to add spectra of one quadrant from multiple observations. In that case the input parameter addexposure need to be set to yes.
To generate spectra for user specified time intervals, there is provision to provide user GTI while generating final GTI for analysis. The user gti file is a text file with two columns in each row corresponding to start and end times of a good time interval. The times have to be in AstroSat time, which is number of UTC seconds from 2010-Jan-1 0 UTC. As an example we will generate spectra for two time intervals of the observation ID 2432 in the example given in Section 1.
We will generate spectra for two time intervals as listed below as an example.
1. 277202800 – 277208000
2. 277209000 – 277213800
Two text files with these user GTI are to be created first, named as ‘usergti_trange1.txt’ and ‘usergti_trange2.txt’ in this example. Then, we execute cztpipeline twice with these user GTI inputs as given below. Also note that we use suffix option to have different names for the output files for each time range.
For the first interval:
$ cztpipeline suffix='trange1'
--------------------------------------------------------------------------------------------------
ASTROSAT CZTI PIPELINE
Version: V3.0 December 2022
--------------------------------------------------------------------------------------------------
Enter Start Step [2]:
Enter End Step [3]:
Enter path to create L2 pipeline products []:
/home/user/path/data_czti/20181014_C04_007T02_9000002432_level2/
Enter full path of user-gtifile [-]: usergti_trange1.txt
Enter output type <lc/spec/both>(for cztbindata) [lc]: spec
Apply maskweights (YES/NO)(for cztbindata) [Yes]: Yes
Enter RA of source in decimal degrees [0]: 83.63308
Enter DEC of source in decimal degrees [0]: 22.01446
Overwrite existing file?(YES/NO): [YES]:
Now, run for the second interval:
$ cztpipeline suffix='trange2'
--------------------------------------------------------------------------------------------------
ASTROSAT CZTI PIPELINE
Version: V3.0 December 2022
--------------------------------------------------------------------------------------------------
Enter Start Step [2]:
Enter End Step [3]:
Enter path to create L2 pipeline products []:
/home/user/path/data_czti/20181014_C04_007T02_9000002432_level2/
Enter full path of user-gtifile [-]: usergti_trange2.txt
Enter output type <lc/spec/both>(for cztbindata) [lc]: spec
Apply maskweights (YES/NO)(for cztbindata) [Yes]: Yes
Enter RA of source in decimal degrees [0]: 83.63308
Enter DEC of source in decimal degrees [0]: 22.01446
Overwrite existing file?(YES/NO): [YES]:
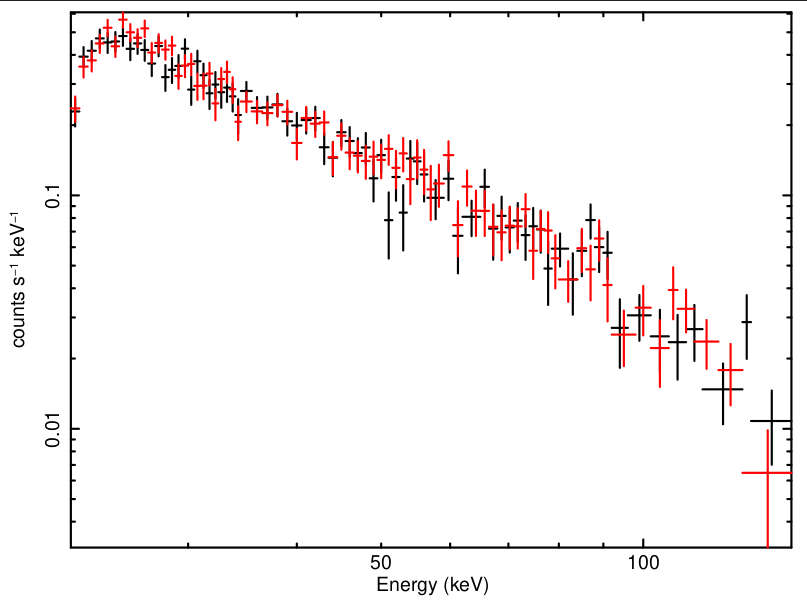
This would produce spectra and responses for the two time ranges within the level-2 directory having names with suffixes trange1 and trange2. Figure 2 shows quadrant 0 spectra for the two time ranges.
 |
|---|
| Figure 2: CZTI crab spectra with quadrant 0 of CZTI for two intervals. |